Decoding X-ray Hand Bones | Semantic Segmentation
1. Introduction
Bone Segmentation is one of the most important applications in Artificial Intelligence, using deep learning technologies to find and segment individual bone.
Thanks to boostcamp AI Tech, our team can work on a project to segment and analyze hand bones using semantic segmentation models for a medical image dataset, generally hard to obtain.
As a team leader on this project, I contributed to managing our project schedule, creating a visualization tool to analyze dataset or experiment results, conducting model experiment based on relevant libraries, and carrying out ensemble to improve the final result of our models.
2. Exploratory Data Analysis (EDA)
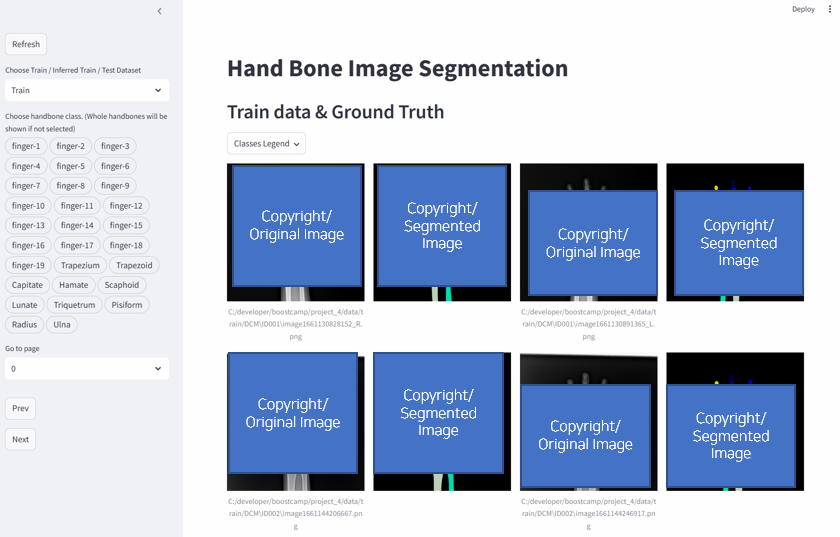
2.1. Visualization Tool
A visualization tool is introduced using Streamlit, an open-source framework for AI/ML engineers. To analyze the image data properly, we needed features as follows.
- Compare (image vs label, image vs model result, model result vs label)
- Toggle the visibility of labels by class
- Upload experiment result (in csv format)
Considering all the requirements above, I made web demo for data(images & labels) visualization as below. You can find the code here.
2.2. What we found
Through our web visualizer, we found a few things to consider in this project. First, the pixels on the wrist bone are overlapped compared to the finger bones. It would be effective to choose a model that works for medical images. Next, some bones such as fingertip or wrist are too small to identify with naked eyes. Enlarging the image could be a solution. Also, small number of data is also one of the limitations to improve the performance of our model.
3. Model Experiments
3.1. Segmentation models
Segmentation has already been developed as a relatively traditional deep learning technology in AI, which changes so quickly. Fortunately, there is a library, called Segmentation Models, which supports various segmentation models and encoders. To conduct experiments quickly, we adopted this library. The interface for model application looks like as follows.
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
import segmentation_models_pytorch as smp
class SmpModel:
def __init__(self, model_config, num_classes):
self.arch = model_config['arch']
self.encoder_name = model_config['encoder_name']
self.encoder_weights = model_config['encoder_weights']
self.in_channels = model_config['in_channels']
self.num_classes = num_classes
def get_model(self):
self.model = smp.create_model(
arch = self.arch,
encoder_name = self.encoder_name,
encoder_weights = self.encoder_weights,
in_channels = self.in_channels,
classes = self.num_classes,
)
return self.model
Considering the characteristics of our dataset, I experimented with medical-originated models (UNet, MANet) or the latest models (UPerNet, SegFormer).
- UNet Series
| Model | Backbone | Dice |
|---|---|---|
| UNet | ResNet50 | 0.9474 |
| UNet++ | ResNet50 | 0.9538 |
| UNEt++ | ResNet101 | 0.9517 |
| UNEt++ | Efficientnet-b5 | 0.9511 |
| UNEt++ | GerNet-L | 0.9513 |
- MANet : Due to limited time, the training was executed with smaller image size(512 ➡️ 256) and less epochs (100 ➡️ 50)
| Model | Backbone | Dice |
|---|---|---|
| UNet++ | ResNet50 | 0.9101 |
| MANet | ResNet50 | 0.8989 |
| MANet | Res2Net50_26w_4s | 0.8957 |
| MANet | ResNest50d_4s2x40d | 0.9026 |
- UPerNet
| Model | Backbone | Dice |
|---|---|---|
| UperNet | ResNet101 | 0.9479 |
| UperNet | swin transformer | 0.9501 |
- SegFormer
| Model | Backbone | Dice |
|---|---|---|
| SegFormer | mit-b0 | 0.9610 |
3.2. Augmentation
Furthermore, we performed off-line augmentation by cropping and enlarging small bones or wrist in order to solve small bone and overlapping problems.
- Size
As the below table shows, there was a significant performance improvement when the size increased. However, it was impossible to enlarge image size over 1024 due to memory limitations.
| Image Size | Dice |
|---|---|
| 256 x 256 | 0.8431 |
| 512 x 512 | 0.8575 |
| 1024 x 1024 | 0.9644 |
- Off-line augmentation
Through cropping wrist or/and fingertip images, we added more images into our dataset. The result shows that the performance was improved, which seems to be due to 1) an increase in the amount of data itself, 2) additional learning for the part that lacked training.
| Class | Original | Wrist | Fingertip | Wrist + Fingertip |
|---|---|---|---|---|
| Lunate | 0.9103 | 0.9366 | 0.9340 | 0.9316 |
| Trapezoid | 0.8705 | 0.8951 | 0.8917 | 0.9005 |
| Pisiform | 0.8232 | 0.8750 | 0.8689 | 0.8864 |
| Trapezium | 0.9110 | 0.9221 | 0.9082 | 0.9178 |
| finger-16 | 0.8564 | 0.9060 | 0.9054 | 0.9002 |
4. Ensemble
In data science, ensemble learning is a technique that combines multiple models to make more accurate prediction. Hard voting-based ensemble was introduced to improve our model performance. Combining the prediction results of several models for each class pixels, it is reflected in the final result when vote result exceed threshold. The way how I coded is as follows.
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
import os
import numpy as np
import pandas as pd
from tqdm import tqdm
def decode_rle_to_mask(rle, height, width):
s = rle.split()
starts, lengths = [np.asarray(x, dtype=int) for x in (s[0:][::2], s[1:][::2])]
starts -= 1
ends = starts + lengths
img = np.zeros(height * width, dtype=np.uint8)
for lo, hi in zip(starts, ends):
img[lo:hi] = 1
return img.reshape(height, width)
def encode_mask_to_rle(mask):
pixels = mask.flatten()
pixels = np.concatenate([[0], pixels, [0]])
runs = np.where(pixels[1:] != pixels[:-1])[0] + 1
runs[1::2] -= runs[::2]
return ' '.join(str(x) for x in runs)
def csv_ensemble(csv_paths, save_dir, threshold):
csv_data = []
for path in csv_paths:
data = pd.read_csv(path)
csv_data.append(data)
file_num = len(csv_data)
filename_and_class = []
rles = []
print(f"Model Number : {file_num}, threshold: {threshold}")
pivot_df = csv_data[0]
for _, row in tqdm(pivot_df.iterrows(), total=pivot_df.shape[0]):
img = row['image_name']
cls = row['class']
model_rles = []
for data in csv_data:
rle_value = data[(data['image_name'] == img) & (data['class'] == cls)]['rle'].values
if len(rle_value) == 0 or pd.isna(rle_value[0]):
print(f"No RLE Data : Image {img}, Class {cls}")
model_rles.append(np.zeros((2048, 2048), dtype=np.uint8))
continue
model_rles.append(decode_rle_to_mask(str(rle_value[0]), 2048, 2048))
image = np.zeros((2048,2048))
for model in model_rles:
image += model
image[image <= threshold] = 0
image[image > threshold] = 1
result_image = image
rles.append(encode_mask_to_rle(result_image))
filename_and_class.append(f"{cls}_{img}")
classes, filename = zip(*[x.split("_") for x in filename_and_class])
image_name = [os.path.basename(f) for f in filename]
df = pd.DataFrame({
"image_name": image_name,
"class": classes,
"rle": rles,
})
df.to_csv(save_dir, index=False)
5. Conclusion
5.1. Result
Based on the experiment results, we concluded and trained our final models for 100 epochs.
- Unet++ : Backbone(ResNet50), Image Size(1024)
- DeepLabV3 : Backbone(EfficientNet-b8), Image Size(1024)
- UperNet : Backbone(Swin Transformer), Image Size(1536)
- SegFormer : Backbone(mit-b0), Image Size(1024)
With the results, we could derive the best performance compared to the baseline. (3.4%🔺)
| Model | Threshold | Dice |
|---|---|---|
| Ensemble (w/o Best) | 1 | 0.9676 |
| Ensemble (w/o Best) | 2 | 0.9689 |
| Ensemble (w/ Best) | 1 | 0.9696 |
5.2. Copyright and Code
This project is a part of a project held at boostcamp AI Tech, managed by NAVER Connect Foundation. The dataset is only used for boostcamp education and not allowed to take outside, so this post includes very limited image. Furthermore, it is available to find more detailed code for this project in my GitHub.